Next-Generation Sequencing, Bioinformatics, and AI
We are back at The Lab...Unlocking Insights, Sparking Curiosity, and Transforming Your Business.
🌷🧪 Spring is in the air, and it's the perfect time to freshen things up at The Lab!
Each month, I'll share some of the latest trends and insights shaping biotech and digital health. I'll also give you a view into the bleeding-edge techniques I use to help businesses make data-driven decisions.
In addition to industry insights, I'll also be sharing a thought-provoking read that I've enjoyed each month.
From time to time, I’ll still share long-form write-ups to explore key topics in greater detail.
📈 Trends & Insights: The Convergence of NGS, AI, and Bioinformatics
Next-generation sequencing (NGS) has revolutionized the field of genomics, enabling researchers to generate vast amounts of genetic data at an unprecedented scale. However, the sheer volume and complexity of this data pose significant challenges for analysis and interpretation. The integration of AI/ML into bioinformatics workflows is becoming a powerful asset to unlocking the full potential of NGS data.
The Role of AI and ML in NGS Data Analysis
AI/ML is increasingly being applied to various stages of the NGS data analysis pipeline, from sequence alignment to variant annotation and beyond. Some key applications include:
Sequence Alignment: optimize the process of aligning sequences to a reference genome by finding the best matching sequences and adjusting for errors, which is crucial for identifying variations and mutations.
Variant Annotation: With the ability to analyze large numbers of variants in NGS data, AI/ML can help identify the most relevant ones associated with diseases or functional changes.
Transcriptomics Analysis: predict gene functions by analyzing patterns in gene expression data generated by NGS.
Tool Development: AI/ML is being used to develop new NGS analysis tools and methods, such as algorithms that predict assay performance or identify new ways to analyze the data more accurately and efficiently.
Error Detection and Correction: Predictive models trained on historical data can be used to detect and correct errors in the NGS analysis process.
Data Visualization: AI/ML can summarize and visualize NGS data to provide meaningful insights, distilling the large amount of information into the most relevant points.
Report Generation: AI-powered natural language generation (NLG) can automate the creation of human-understandable reports from raw NGS data.
An example for AI and variant calling is Google Brain's DeepVariant is a deep learning-based variant caller that uses convolutional neural networks (CNNs) to analyze NGS data. It learns to identify variants by training on large datasets of sequencing reads aligned to reference genomes. DeepVariant has demonstrated superior accuracy compared to traditional variant calling methods, particularly in challenging regions of the genome.
Illumina's DRAGEN (Dynamic Read Analysis for GENomics) Bio-IT Platform uses hardware-accelerated AI algorithms to speed up NGS data analysis. It can perform sequence alignment, variant calling, and other computationally intensive tasks up to 50 times faster than traditional methods. This enables researchers to process and analyze NGS data more efficiently, accelerating the pace of genomic research.
The ability of AI/ML to quickly and accurately process vast quantities of complex data makes these technologies essential for modern NGS analysis. Deep learning, in particular, is advantageous for identifying genetic patterns in the exponentially growing genomic datasets.
The Impact of NLP on Biomedical Research
Natural language processing (NLP), a subfield of AI, is also making significant strides in the biomedical domain. Language-based AI has advanced rapidly in recent years, changing notions of what the technology can do in areas like writing, coding, and reasoning. In the context of NGS and bioinformatics, NLP tools can help with tasks such as extracting information from scientific literature and clinical notes, facilitating knowledge discovery and decision support.
Examples of how NLP tools are being used in support of NGS:
Literature mining: NLP can be used to extract relevant information from scientific literature, such as gene-disease associations, protein-protein interactions, and drug-target relationships. This can help researchers identify potential targets for drug repurposing or discover new biological insights from existing knowledge.
Clinical note analysis: NLP can be applied to electronic health records (EHRs) and clinical notes to extract valuable information, such as patient phenotypes, treatment outcomes, and adverse events. This data can be integrated with NGS data to identify genetic variants associated with specific clinical outcomes or to stratify patients for precision medicine applications.
Automated report generation: NLP can be used to generate human-readable reports from raw NGS data, summarizing key findings and highlighting clinically relevant variants. This can save time and reduce errors in manual interpretation of NGS results.
The Future of NGS and AI
As NGS technologies continue to advance and datasets grow, AI will become increasingly critical for unlocking the full potential of genomic data. The convergence of NGS, AI/ML, and bioinformatics is enabling faster, more accurate, and automated analysis of the huge volumes of data generated by NGS. This is accelerating research and unlocking new biological insights and clinical applications, from understanding disease mechanisms to developing personalized therapies.
However, there are also challenges to be addressed, such as the need for standardized data formats, interoperability between tools, and the integration of domain knowledge into AI/ML models. Collaboration between experts in genomics, bioinformatics, and AI will be key to overcoming these hurdles and realizing the full promise of these technologies.
In conclusion, the intersection of NGS, AI, ML, and bioinformatics represents a powerful and transformative force in genomic research. As these fields continue to evolve and intertwine, we can expect to see even more breakthroughs and innovations in the years to come, ultimately leading to better understanding of biology and improved human health.
The Patent Landscape
An analysis of the patent landscape reveals a growing interest and investment in the convergence of NGS, AI/ML, and bioinformatics. There is an increasing number of patent filings over the years, which suggests that companies and researchers recognize the potential of these technologies to transform drug discovery, precision medicine, and other areas of the life sciences industry.
The bar chart below (Figure-1) shows a general upward trend in patent filings in this area over the years, with "Bioinformatics Analysis" being the most frequently occurring term, indicating a strong focus on the computational analysis of biological data within the patent filings.
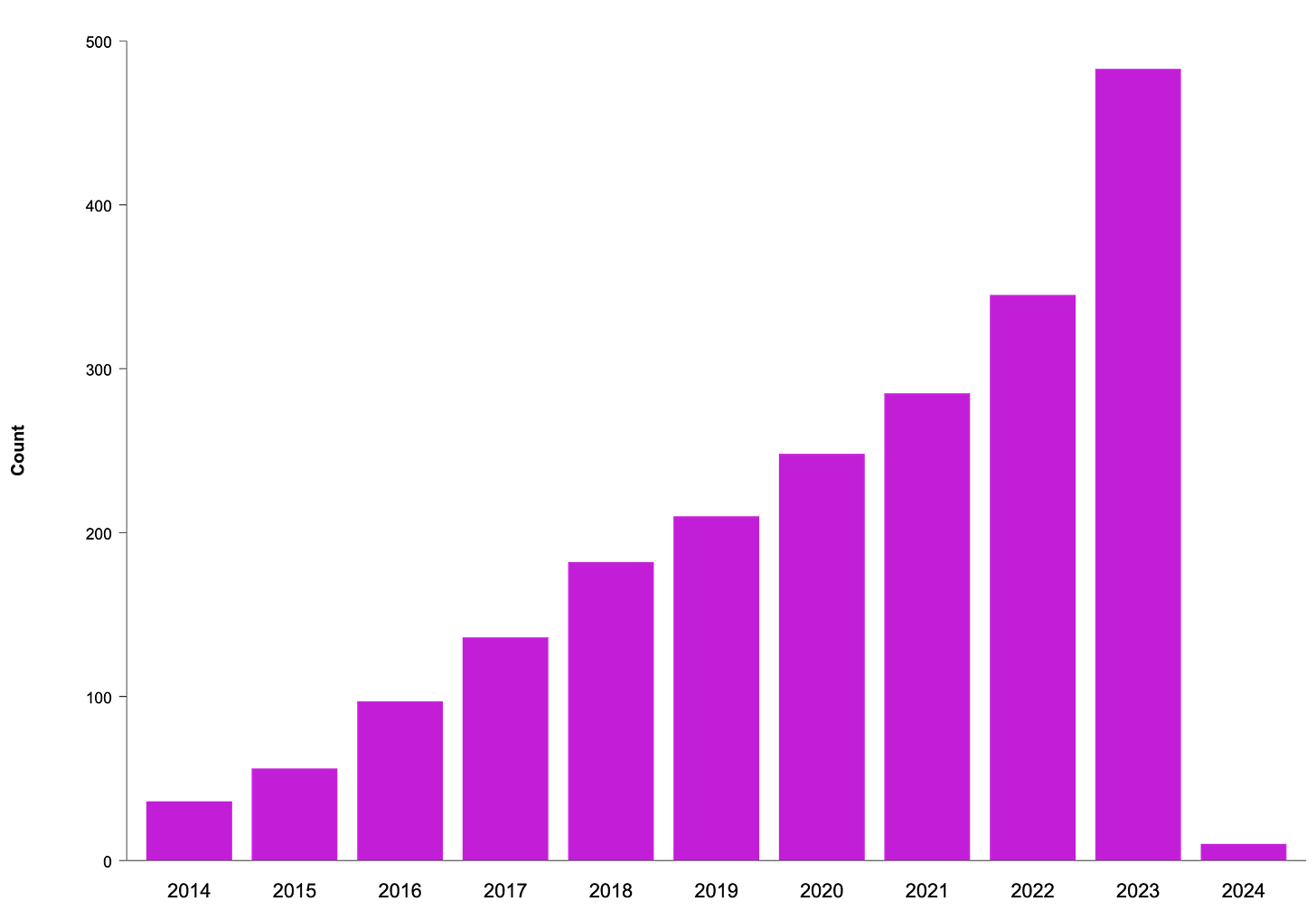
The global nature of the patent filings, with leading contributions from countries like China, the United States, India, and South Korea, indicates a competitive and dynamic landscape. Companies and research institutions around the world are racing to develop and protect their innovations in this space, which could lead to a rapid pace of technological advancement and new market opportunities.
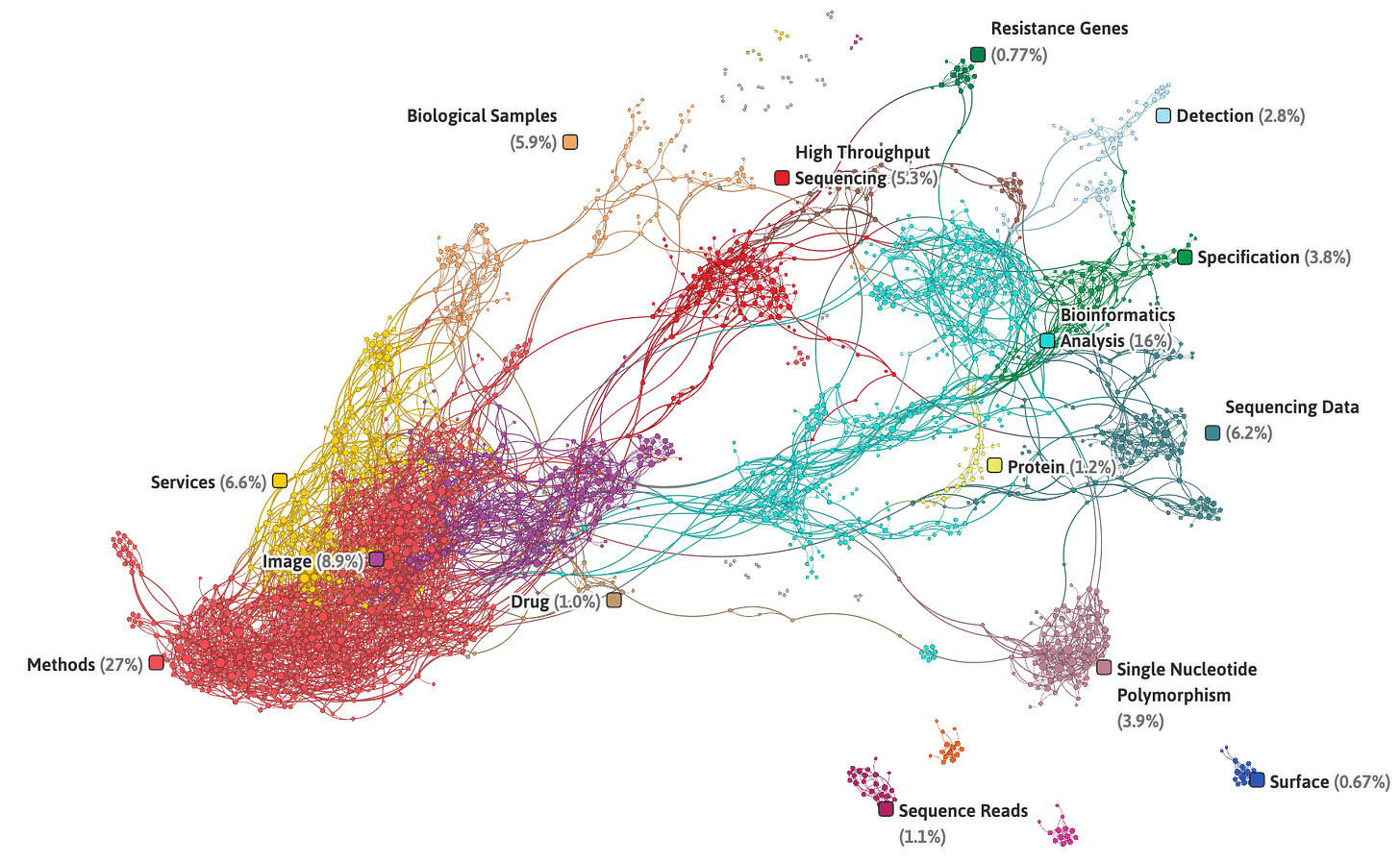
The network below (Figure-2) provides a visual representation of patents related to the fields of NGS, AI/ML, and bioinformatics over the past decade.
The central nodes represent core areas such as High Throughput Sequencing (5.3%), Bioinformatics Analysis (1.6%), and Resistance Genes (0.7%). These are the major focus areas for patenting activity. Sequencing Data (6.2%), Protein (1.2%), and Single Nucleotide Polymorphism (3.9%) are the focus of methods and applications patents related to these area.
Nodes for Detection (2.8%), Specification (3.8%), and Surface (0.6%) technologies suggest patented innovations in sample preparation, sequencing, and analysis techniques.
The Drug (1.0%) and Image (0.9%) nodes indicate patented applications of NGS, bioinformatics, and AI in drug discovery and medical imaging analysis.
The complex interconnections between nodes highlight the interdisciplinary nature of these fields and potential cross-utilization of methods and data across different applications.
Overall, these trends suggest a robust and global effort to innovate and secure intellectual property in these critical areas of research and development, which are essential for the progress of genomics, personalized medicine, and the broader life sciences industry.
📚Good Read of the Month
“Drug Repurposing Strategies, Challenges and Successes” By Megan Sperry, PhD & Prof. Donald E. Ingber, MD, PhD
This article provides a look inside the process of drug repurposing that can empower and inform new product features and strategies for pharma partners. Whether you’re offering any products or services in this area or work in the pharma industry, you’ll find this article interesting!
Background
The drug discovery process is riddled with failures so drug repurposing can be a smart strategy to rescue the investment.
Drug repurposing, also known as drug repositioning, is the process of identifying new therapeutic uses for existing drugs that have been approved, failed in clinical trials, or withdrawn from the market due to safety concerns. This approach involves finding new indications for drugs beyond their original purpose, often leveraging common biological targets shared by different diseases. At this stage, the pharmaceutical industry relies on technology providers and CROs to help with AI-powered modeling, advanced analytics, and testing.
Quick Insights:
Repurposed drugs can be approved faster (within 3-12 years) and at about half the cost compared to traditional drug development. Around 30% of repurposing efforts result in an approved product, compared to about 10% for new drug applications.
Key barriers to successful drug repurposing include inadequate resources, lack of access to trial data and information on abandoned compounds, and challenges in negotiating intellectual property (IP) agreements.
Technical challenges in drug repurposing include data volume and hygiene issues, data heterogeneity, and the need for advanced computational methods.
As always, thank you for supporting The Lab!
Reach out with your thoughts, ideas, and questions. Find me on LinkedIn or shoot me an email.
Cheers,
Mida